XPERANTO
Research background
DNA microarray is a representative of high-throughput biomedical
technologies that allow monitoring of gene expression for thousands
of genes in parallel. DNA microarray experiment generates enormous
amounts of data and they are meaningful only in the context of a
detailed description of microarrays, biomaterials, and conditions
under which they were generated. As microarrays are increasingly
used in a variety of biomedical research, the systematic management
of diverse and large amount of gene expression data has received
much attention. In particular, structured data storage and use of
controlled terms with a well-being meaning are important for easy
access and efficient use of the data.
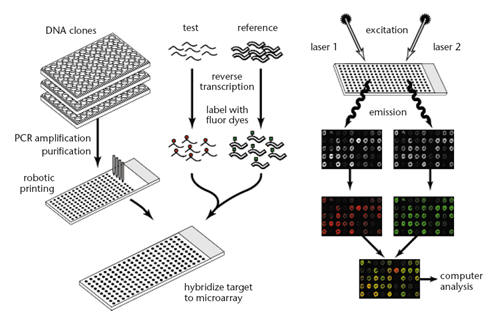
Experiment
procedure and data format of DNA microarray
figure from Nat. Genet. 21:10-14 (1999) and 29:365-71 (2001)
Purpose
With the growing needs of data sharing, Microarray Gene Expression
Data (MGED) society has established common standards for microarray
data. MIAME (Minimum Information About a Microarray Experiment) is a
data annotation standard to specify the minimum information that
should be reported about a microarray experiment. MAGE (MicroArray
Gene Expression) standards – an object model (MAGE-OM) and an XML
based language (MAGE-ML) – have been developed for the
representation of microarray expression data, facilitating the
exchange of microarray information between different systems. MGED
Ontology group defines sets of common terms and annotation rules for
microarray experiments. Although there are a number of microarray
databases in existence, it is not easy to find the database meeting
the specialized requirements of individual institutions or groups
for local data archiving, analysis and sharing. The complexity of
microarray data requires an integrated environment that does much of
the data storage, visualization, analysis, and transfer. Based on
this, we present Xperanto, meaning eXpressionist¨s Esperanto in XML,
which is a microarray information system for microarray studies.

A schematic representation of six
components of a microarray experiment
figure from Nat. Genet. 29:365-71 (2001)
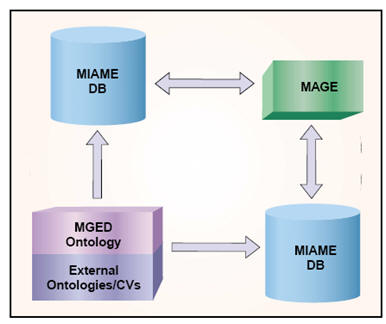
Relationships among
the different MGED initiatives
figure from Nat. Genet. 32:469-473 (2002)
Methods
- Database design
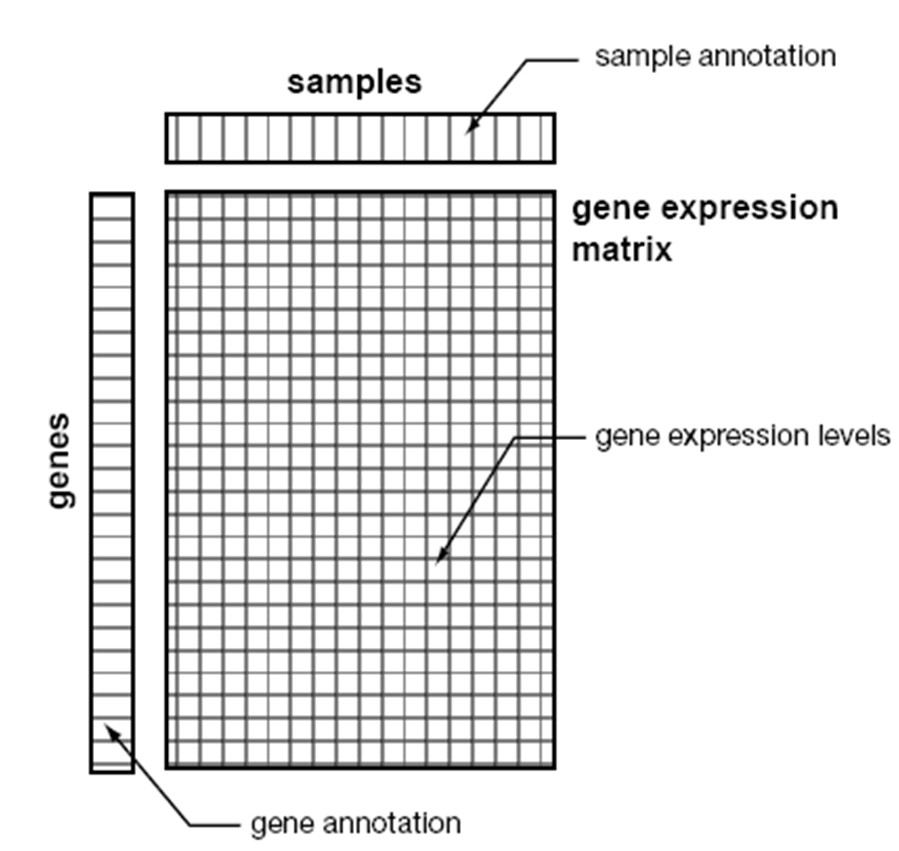
Xperanto database is basically designed to collect all contents defined by the MIAME standard and meet the specific requirements of laboratories. The database is comprised of six components: experiment, hybridization, biomaterial, array, protocol, and gene expression data (experimentally measured and computationally processed). By experiment we understand a set of one or more hybridizations that are in some way related, often corresponding to a particular publication. Values for data items come from look-up table that manages controlled terms from the MGED Ontology (http://www.mged.org/ontology/). To separate the data utilization according to research group, we implemented a security system allowing each investigator to securely manage their own microarray data and analysis results.
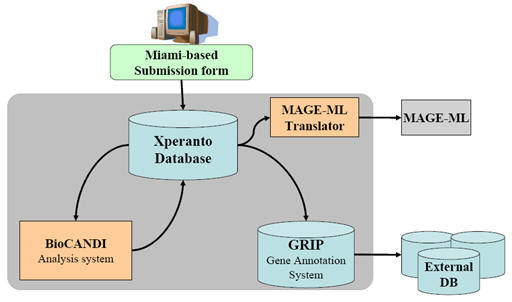
System architecture of Xperanto
- Developmental environment
For the efficient development of the system, we analyzed the user's requirements so as to define the functional and behavioral aspects of the system. All the results were well documented to describe the various views of the microarray information system, and the document helps the communication among a wide range of the scientists involved in the production and analysis of microarray data.
- User interface
Xperanto provides easy use for researchers by modeling a natural workflow in the microarray experiment. It also aims at encouraging more complete and accurate recordings with the structured data entry system such as drop-down lists of the MGED Ontology terms. Submissions are validated syntactically so that the records are organized correctly. The experimental data and their biological and statistical context are intra-linked on the system, thereby increasing the accessibility to the associated records. To accept various types of quantitative data from image-analysis programs, utility programs are available for modifying their format in system-defined way.
- Efficient data storage and management

Workflows that lead up to a hybridized microarray
(Rectangles represent physical things, diamonds represent events, and rounded rectangles represent methods.) - Xperanto-CGH
The mechanism of cancer progression involves chromosomal aberrations, including deletion of tumor suppressor genes and amplification of oncogenes. Microarray-based comparative genomic hybridization (array-CGH) is uniquely well suited for high-resolution detection of DNA copy number aberrations.
Xperanto-CGH is an integrated system for array CGH data
management and analysis.
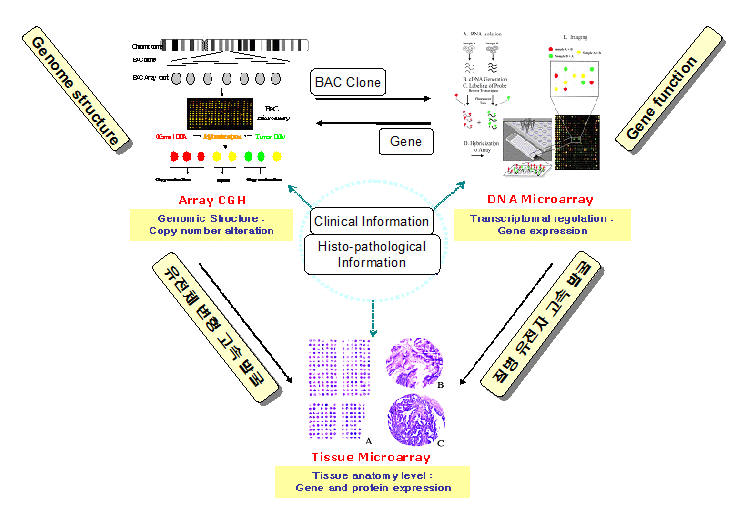
Data storage system for DNA microarray, array CGH, and tissue microarray
- Xperanto-Tox
For supporting toxicologist who comes up against toxicogenomic data flood, we propose novel toxicogenomics knowledgebase system, Xperanto-Tox. Xperanto-Tox is an integrated system for toxicogenomic data management and analysis. It is composed of three distinct but closely connected parts; Data storage system, Data analysis system, Data annotation system.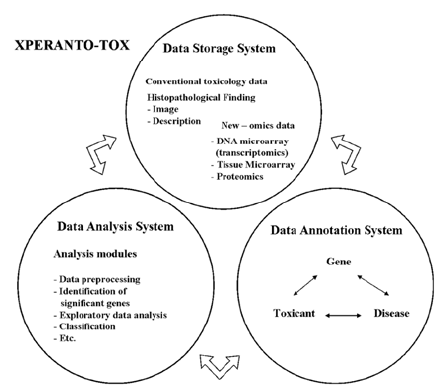
The composition of whole system
- Xperanto-SNP
We developed Xperanto-SNP for integrated data management and analysis using user-friendly web-based interface. Xperanto-SNP provides an integrated environment for management and analysis by linking the computational tools and rich sources of biological annotation to get meaningful knowledge. Xperanto-SNP enables a fast and efficient management of vast amounts of data, and serves as a communication channel among multiple researchers within an emerging interdisciplinary field.
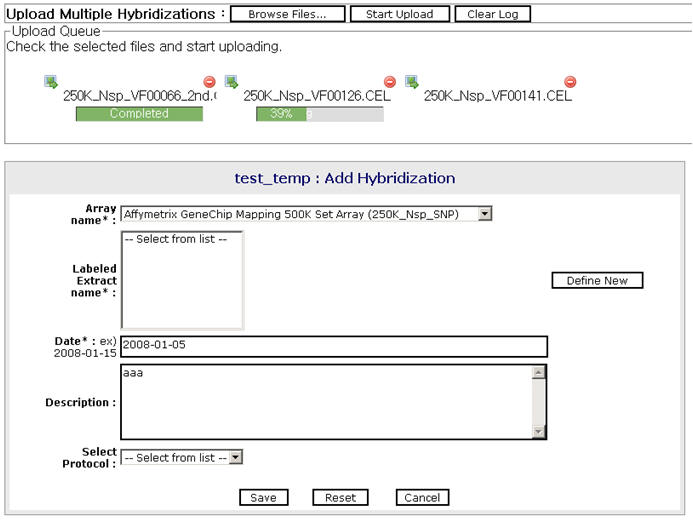
SNP chip data storage