| 일정 | 주제 | 발표자 | |
|---|---|---|---|
| 08:00 ~ 10:30 | 집합 및 출발 | ||
| 10:30~10:40 | 개회 | ||
| 10:40 ~ 11:40 | Session 1 | ||
| 10:40~11:00 | Bayesian inference of genetic models of complex diseases | ||
| 11:00~11:20 | Bladder Cancer Cell lines and its Chemo-resistance profiles | ||
| 11:20~11:40 | 휴 식 | ||
| 11:40 ~ 12:20 | Session 2 | ||
| 11:40~12:00 | Genomic characterization of drug perturbation sensitivity | ||
| 12:00~12:20 | Identifying disease variants in genotypic association | ||
| 12:20~13:20 | 중 식 | ||
| 13:20 ~ 14:20 | Session 3 | ||
| 13:20~13:40 | Aging Pattern Clustering - Gender Difference | ||
| 13:40~14:00 | Early detection of pharmacovigilance adverse signals using controlled vocabularies | ||
| 14:00~14:20 | MELLO: Medical Life-Log Ontology | ||
| 14:20~14:40 | 휴 식 | ||
| 14:40~15:40 | Session 4 | ||
| 14:40~15:00 | SNPChase : correcting errors from diverse SNV databases for SNVs | ||
| 15:00~15:20 | PhenoRanker: Personal genome sequence based phenotype ranking algorithm for potential risk prediction� | ||
| 15:20~15:40 | PharmRank2:Personal genome sequence-based pharmacogenomic drug ranking with biomedical knowledge boosting | ||
| 15:40~16:00 | 휴 식 | ||
| 16:00~16:30 | Session 5 | ||
| 16:00~16:30 | Health avatar platform 시연 | ||
| 16:30~16:40 | 폐 회 | ||
| 16:40~18:30 | 짐 정리 및 개인활동 | ||
| 19:00~ | 저녁식사 | ||
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
2013 Winter Retreat
주최: 서울대학교 의과대학 정보의학실
장 소: 대명 비발디파크, (http://www.daemyungresort.com/vp) 강원도 홍천군 서면
시 간: 2013년 2월 4일(월) - 6일 (수)
Program - Day 1| 일정 | 주제 | 발표자 | |
|---|---|---|---|
| 08:30 ~ 10:00 | 집합 및 출발 | ||
| 10:00~10:20 | 개회 | ||
| Session 1 | Microarray Analysis & Functional Genomics | ||
| 10:20~10:50 | Integrative Analysis of microRNA-target Interactions with Clinical Outcome | ||
| 10:50~11:20 | Functional Characterization of Yeast DNA Motif Clusters using Multiple Annotation Methods | ||
| 11:20~11:50 | Knowledge bootstrapping: A graph-based integration with multi-omics data and genomic knowledge | ||
| 11:50~13:30 | 중 식 | ||
| Session 2 | NGS Data Analysis & Genome Interpretation (Part I) | ||
| 13:00~13:30 | Signature of Nephrotic Syndrome: Whole Transcriptome Sequencing of Peripheral Mononuclear Cells from Pediatric Idiopathic Nephrotic Syndrome | ||
| 13:30~14:00 | Characterizing gene expression in lung tissue of COPD subjects using RNA-seq | ||
| 14:00~14:20 | 휴 식 | ||
| 14:20~14:50 | Genomancer : A secure, modular and distributed system for interpreting personal genome on mobile smartphone | ||
| 14:50~15:20 | CREE a Tool for Comparison and Retrieving of RNA-editing Sites from RNA-seq Data | ||
| 15:20~15:40 | 휴 식 | ||
| Session 3 | Medical Informatics (Part I) | ||
| 15:40~16:10 | Metadata Registry based integration and transformation between openEHR archetype and HL7 template | ||
| 16:10~16:40 | CDISC Transformer: metadata driven semi-automatic transformation of clinical trial data to CDISC ODM | ||
| 16:40~17:40 | 종 료 | ||
| 일정 | 주제 | 발표자 | |
|---|---|---|---|
| Session 1 | NGS Data Analysis & Genome Interpretation (Part II) | ||
| 09:00~09:30 | Exome sequencing of Acute Myeloid Leukemia, M2 subtype | ||
| 09:30~10:00 | MedCassandra: Drug and ADR ranking forecast system based on personal genomic variation | ||
| 10:00~10:30 | Predicting disease predisposition patterns of the personal genome | ||
| 10:30~10:50 | 휴 식 | ||
| Session 2 | Medical Informatics (Part II) | ||
| 10:50~11:20 | HealthPro: Designing Search Strategies for Retrieving Data from Clinical Data Warehouse | ||
| 11:20~11:50 | Rapid identification of adverse drug reaction using controlled vocabularies | ||
| 11:50~12:00 | 폐 회 | ||
2012 Spring Retreat
주최: 서울대학교 의과대학 정보의학실
장 소: 라 데나 콘도미니엄, (http://www.ladenaresort.com) 강원도 춘천시 삼천동
시 간: 2012년 5월 25일(금), 오전 10시 - 오후 6시
Program| 일정 | 주제 | 발표자 | |
|---|---|---|---|
| 10:30~11:00 | 개회 | ||
| Session 1 | NGS Data Analysis & Genome Interpretation | ||
| 11:00~11:30 | Predicting disease predisposition of personal genome based on disease hierarchy | ||
| 11:30~12:00 | Loss of function gene-set analysis of personal genome using pathway-disease similarity | ||
| 12:00~13:30 | 중식 | ||
| Session 2 | Micoarray Analysis & Functional Genomics | ||
| 13:30~14:00 | Extracting of coordinated patterns of DNA methylation and gene expression in ovarian cancer | ||
| 14:00~14:30 | Graph based integration with heterogeneous genomic data and genomic knowledge | ||
| 14:30~15:00 | GEE : A tool for Gene Expression data Explore | ||
| 15:00~15:30 | Coffee break | ||
| Session 3 | Pharmacogenomics & Clinical Informatics | ||
| 15:30~16:00 | Rapid identification of adverse drug reaction using controlled vocabularies | ||
| 16:00~16:30 | HealthPro: An Integrated Informatics Platform for Clinical Research in Data Warehouse | ||
| 16:30~17:00 | 폐회 | ||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2009 Winter Retreat
일정 안내로 바로가기주최: 서울대학교인간유전체연구소 생명의료정보연구실
2009년 겨울 Retreat은 2009년 12월 30일~31일 용인 에코그린타운에서 개최되었습니다.
발표자료 (ppt and paper (word)) Deadline: 12월 29일 6:00pm
Program| 일정 | 주제 | 발표자 | |
|---|---|---|---|
| 개회 | |||
| 10:30~11:00 | Special keynote | 김주한 교수님 | |
| 11:00~12:00 | Session 1: Biomedical data integration research | ||
| 11:00~11:30 | Ontology-based integration of cDNA and tissue microarray database management system | 송영수 | |
| 11:30~12:00 | A graph-based integration of multidimensional cancer genomics data | 김도균 | |
| 12:00~13:30 | 중식 | ||
| 13:30~15:00 | Session 2: Genome data research | ||
| 13:30~14:00 | Characterizing microRNA regulatory modules using microRNA-mRNA coexpression data | 나영지 | |
| 14:00~14:30 | GO Enrichment analysis of clustered miRNAs by using three hypergeometric distribution | 이수연 | |
| 14:30~15:00 | An improved method to measure the gene similarity using ontological relation of GO terms | 변상재 | |
| 15:00~15:30 | Coffee break | ||
| 15:30~17:00 | Session 3: Biological database issue | ||
| 15:30~16:00 | Finding gene symbol, cross-reference inconsistency from Gene Centric Databases | 박찬희 | |
| 16:00~16:30 | MetaPath: a meta-database for public biological pathway database | 정희준 | |
| 16:30~17:00 | Trends in Biological Pathway-based Analysis | 윤선민 | |
| 17:00~17:30 | 폐회 | ||
Dinner: Barbeque party
2008 Winter Retreat
주최: 서울대학교인간유전체연구소 생명의료정보연구실일정 안내로 바로가기
2008년 겨울 Retreat은 2008년 11월 21일 용산역 대회의실에서 개최되었습니다.
Program
2008 Summer Retreat
주최: 서울대학교인간유전체연구소 생명의료정보연구실일정 안내로 바로가기
행사 사진첩으로 바로가기
2008년 여름 Retreat은 2008년 7월 14일 양평 남한강 별장에서 개최되었습니다.
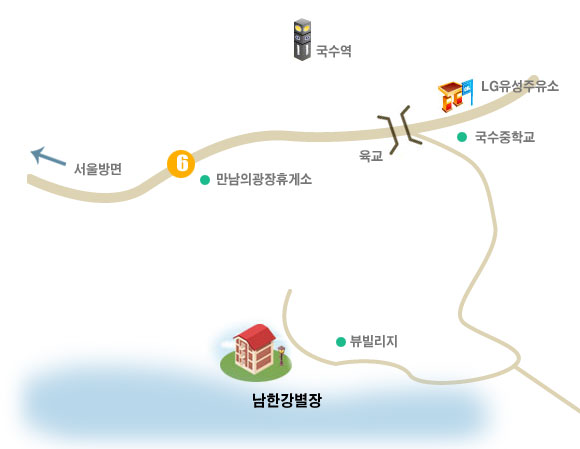
관리인이 상주하는것이 아니므로 답사는 사전에 문의를 주셔야 합니다. [대중교통] 기차 서울의 경우, 청량리역에서 양평행 기차 중 국수역 경유하는 기차를 이용하셔야 합니다. (열차 출발 시간은 역마다 차이가 있습니다. 별도의 확인이 필요합니다) 양평역 하차시 국수리역행 버스로 환승하거나 택시 이용하셔야 합니다(택시비 약 1만원) 버스 상봉역 버스터미널(시외버스이므로 별도의 번호 없이 국수리행 버스 탑승하시면 됩니다. 1시간에 한 두 차례 출발)이나 동서울터미널(2000-1번)에서 버스를 타시고 국수중학교 앞 하차하시면 픽업해 드립니다. - 하차역 : 국수리역,양평 중.고등학교,국수중학교(버스마다 하차역이 다를 수 있습니다) - 하차 후 전화 주시면 픽업해 드립니다. |
Program
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
2008 Summit on Biomedical Informatics
주최: 서울대학교인간유전체연구소 생명의료정보연구실일정 안내로 바로가기
행사 사진첩으로 바로가기
문의사항은 김주한교수님 연구실의 김지훈 (djdoc@snu.ac.kr 02-740-8318)에게 문의해주십시오.
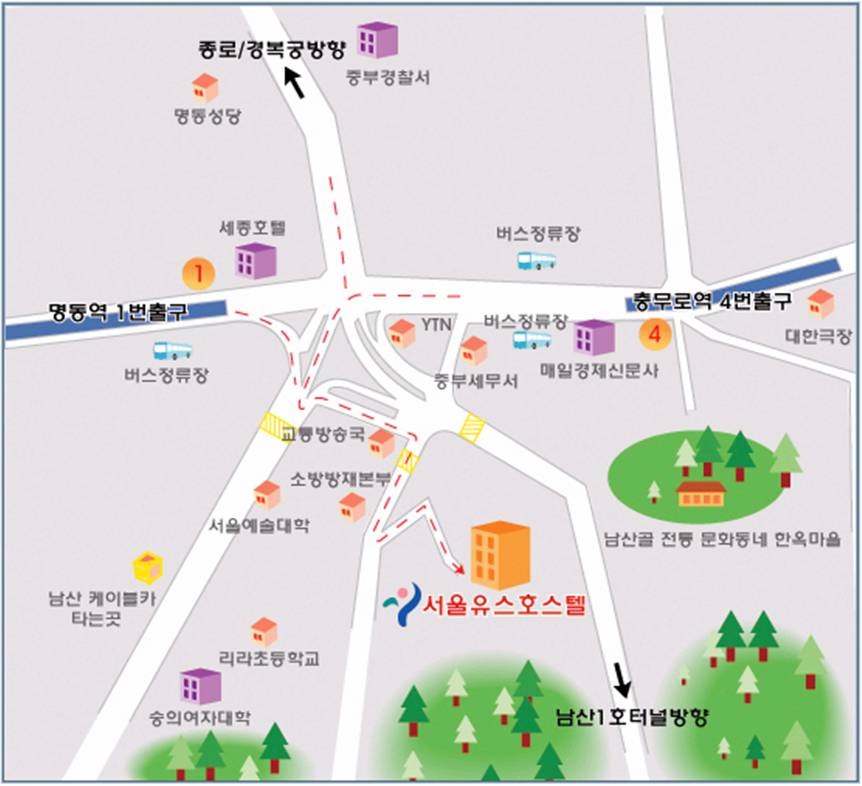
2008년 겨울 첫 번째 세미나는 서울유스호스텔 소회의실 에서 2007년 12월 28일 오전 9시에 시작합니다.
|
Time |
Title |
Abstract |
Presenter |
|---|---|---|---|
|
08:30~09:00 |
Registration |
||
|
09:00~09:50 |
Special keynote |
Professor Ju Han Kim |
|
|
09:50~11:20 |
Session I : Biomedical informatics for disease |
Session chair : Jihun Kim |
|
|
|
The development of clinico-histopathological metadata for cancer-oriented research |
|
Yu Rang Park |
|
|
Pathway classification analysis using BoostKDBC |
|
Sung Bum Cho |
|
|
Feature selection and Rule extraction of time series microarray using GP |
|
Su Yeon Lee |
|
11:20~11:30 |
coffee break |
||
|
11:30~12:30 |
Session II : Young investigator competition |
Session chair : Young Ji NA |
|
|
|
GEOXperanto |
|
Sang Jae Byun |
|
|
Integration of heterogeneous probe-level data |
|
Do Kyoon Kim |
|
12:30~13:30 |
Lunch |
||
|
13:30~15:00 |
Session III : Gene regulation and transcriptomics |
Session chair : Sung Bum Cho |
|
|
|
microRNA regulation depends on the regional effect |
|
Young Ji NA |
|
|
Imporving DEG identification in small sample datasets using GEO data |
|
Mingoo Kim |
|
|
BioCLASS : A semantic comparison of microarray gene expression data |
|
Jihun Kim |
|
15:00~15:10 |
coffee break |
||
|
15:10~16:10 |
Session IV : Database integration |
Session chair : Mingoo Kim |
|
|
|
CeLMAP: integrated pathways map using cellular location information |
|
Hee Joon Chung |
|
|
Integrative analysis in the study of cancer |
|
Chan Hee Park |
|
16:10~16:20 |
Closing |
||
|
16:30~18:30 |
Special program |
||
|
19:00~21:00 |
Reception |
||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|