(This page is still under construction)
MITree
Welcome to bioinformatics tools at CHIP (Children's Hospital Informatics Program).
MITree is a clusterin algorithm based on a straightforward geometric principle. Initially it was designed to be a binary hierarchical clustering algorithm for gene expression analysis. It is well described in Kim JH, Ohno-Machado L, Kohane IS. Unsupervised learning from complex data: the matrix incision tree algorithm. Pac Symp Biocomput 2001;:30-41. We also applied Evolution Strategy (i.e., a genetic algorithm) to the same problem. pdf
Implementation of
MITree is available here with a typical input
data format example (Fisher's iris data set), where data starts from
the 4th column and 2nd row and tab-dilimited ascii text file. We don't
have manual yet, though. MITree-K is a partitional version of MITree.
Many variations of the algorithm is implemented in MITree software.
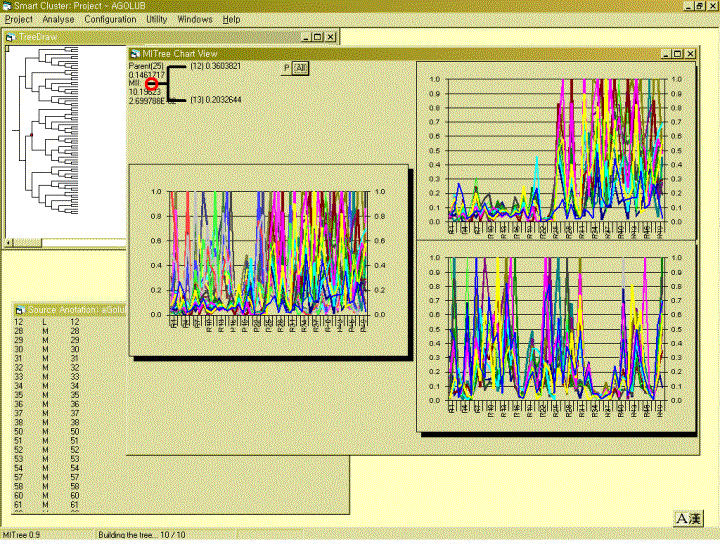
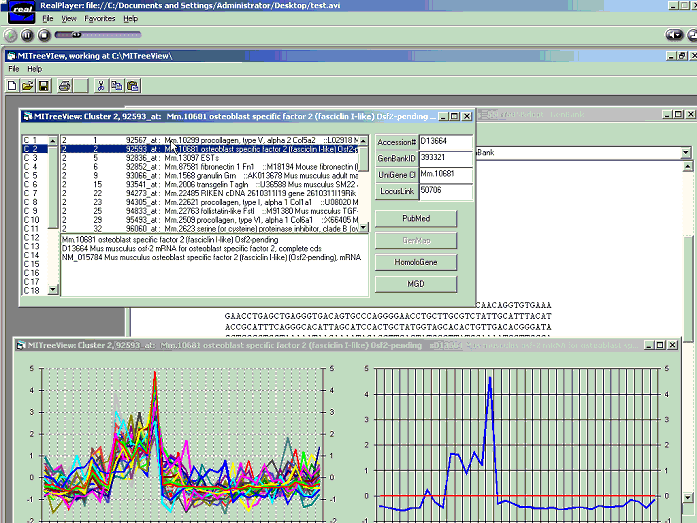
MITreeView is a simple tool to efficiently visualize clusters with related information.
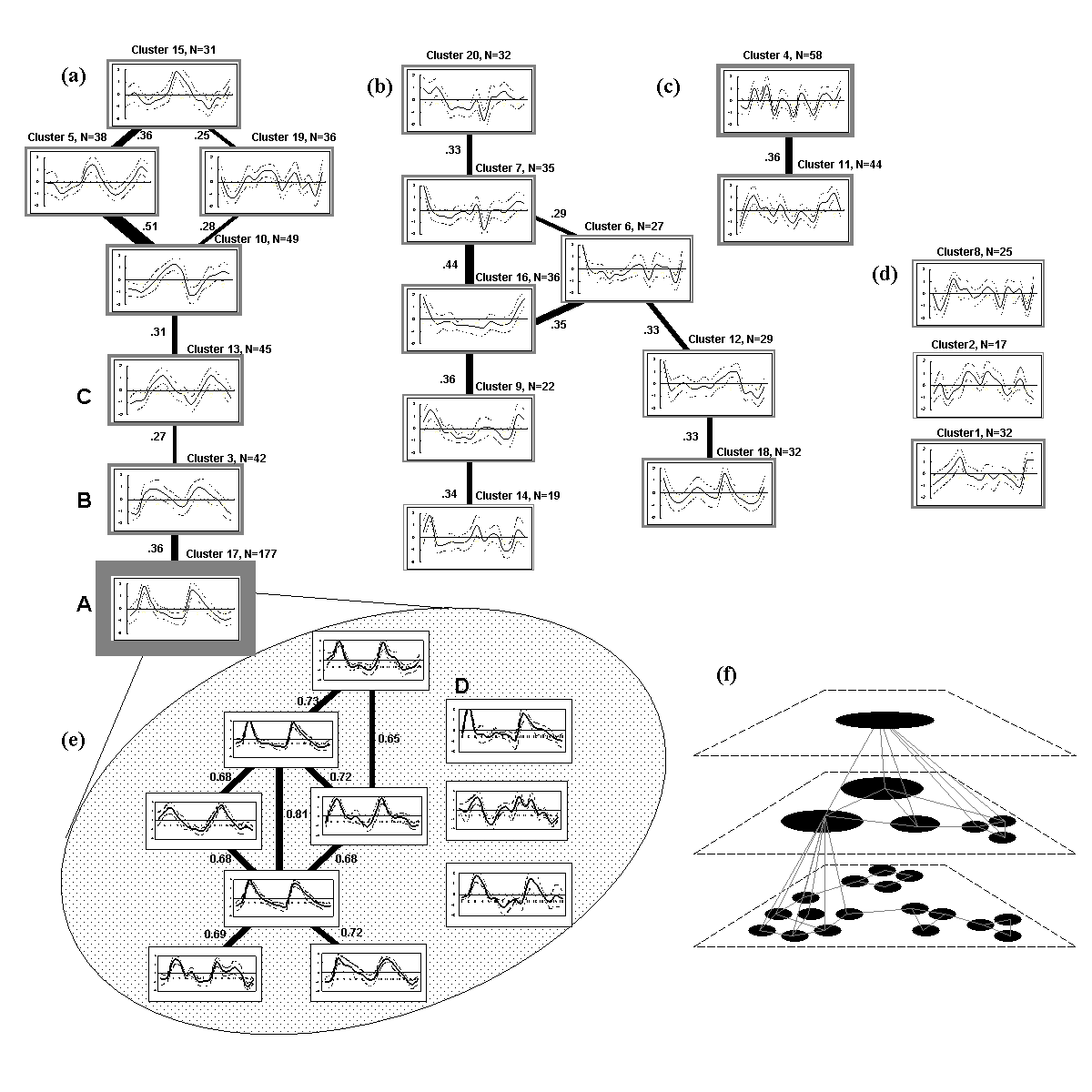
Systematic matrix
decomposition and reconstruction of the clustering results from MITree-K
will return quantiative visualization of the comprehensive clustering structure
as shown below.
Copy rights reserved